Welcome!
Welcome to the Mosig Lab at Jena University Hospital
We are dedicated to advancing human-relevant, immune-competent organ-on-chip systems to unravel host–microbiota interactions and immune responses in infection and inflammation. By integrating patient-derived stem cells, cutting-edge microphysiological models, and AI-assisted analytics, our research bridges basic discovery and translational medicine. Our goal is to reduce animal experiments in line with the 3R principles while creating innovative platforms for personalized therapies and next-generation diagnostics.

RESEARCH
Our research focuses on the development and application of human-relevant, immune-competent organ-on-chip models to study infection, inflammation, and host–microbiota interactions. These models combine patient-derived stem cells, advanced microphysiological systems, and AI-based analytics to enable translation into clinical medicine while adhering to the 3R principles (Replace, Reduce, Refine animal experimentation).
Our mission is to build robust, personalized, and mechanistically grounded in vitro platforms that allow us to:
- uncover fundamental mechanisms of immune–microbiota–pathogen interactions,
- identify new biomarkers and therapeutic targets, and
- contribute to accelerate the development of safer and more effective therapies.
Research Areas
Microbiota & Immunomodulation
We investigate how microbial metabolites, including short-chain fatty acids (SCFAs), secondary bile acids, and amino acid derivatives, affect adaptive immune responses in various contexts such as infection, chronic inflammation, and cancer. Special attention is given to the role of the microbiome in immunotherapies. We are integrating systems biology with AI-driven image and data analysis provided by our partners to create predictive, translational complex human in vitro models that enhance our understanding of principles in host-microbiota interaction under controlled physiologically relevant conditions.
selected projects
- Immune Safety Avatar: nonclinical mimicking of the immune system effects of immunomodulatory therapies (imSAVAR)| funded by the European Commission | 2020–2025
- MPS@NOVA Hub: Mechanisms of chronic diseases and Host-Microbe Interactions with Advanced Microphysiological Systems and Pluripotent Stem Cell Technologies funded by the European Commission | 2024–2027
- Microbiome Interactions in the Respiratory and Gut Epithelium (MIRAGE) | funded by the DFG in the Cluster of Excellence Balance of the Microverse | 2026 – 2029
- Studying the impact of intestinal dysbiosis on respiratory viral infections in a multi-organ model of the gut-lung axis | funded by the DFG in the Cluster of Excellence Balance of the Microverse | 2023 – 2025
Infection Models
Respiratory Infections & Pneumonia
We leverage iPSC-based alveolus- and bronchiolus-on-chip models to study respiratory pathogens, viral–bacterial co-infections, and microbiota-mediated modulation of lung immunity. Our investigations include diagnostics of ventilator-associated pneumonia, cystic fibrosis (CF), and various viral respiratory infections, including RSV and influenza virus. The models of the respiratory tract further serve as a platform for photonic diagnostics and automated testing of novel treatment strategies, such as nanoparticles, for translational applications.
Selected projects:
- Human next-generation mucosal models and assays to accelerate vaccine development (Inno4Vac) | funded by the European Commission | 2021 – 2027
- Exploring the role of pathogen interactions in transitioning of ventilator-associated tracheobronchitis to pneumonia (VATOP) | funded by the DFG | 2026 – 2029
- Modelling cystic fibrosis and associated infections in a bronchiole-on-chip model for drug testing (CF-on-chip) | funded by the Federal Ministry of Economic Affairs and Energy | 2024 – 2026
- Automated organ-on-chip platform for infection research (AutOoC) | funded by the Federal Ministry of Research, Technology and Space | 2025 – 2030
Gastrointestinal Infections
We develop patient-specific intestine-on-chip models to investigate enteric infections caused by pathogens such as Clostridioides difficile, Vibrio cholerae, and Candida albicans. These platforms allow us to dissect key mechanisms of colonization, toxin production, quorum sensing, and dysbiosis. Building on these insights, we evaluate innovative phage and antibiotic therapies, offering strong translational potential for tackling antimicrobial resistance (AMR).
selected projects:
- Human next-generation mucosal models and assays to accelerate vaccine development (Inno4Vac) | funded by the European Commission | 2021 – 2027
- Probing cholera infection on a chip | funded by the DFG in the Cluster of Excellence Balance of the Microverse | 2022 – 2025
- Human infection model for Vibrio cholerae as an alternative to animal testing (VIBRO-3R) | funded by the Federal Ministry of Research, Technology and Space | 2026 – 2029
- Profiling of host-response to life threatening infections | funded by the Federal Ministry of Research, Technology and Space | 2021 – 2027
Parasitology
Together with partners in polymer chemistry (Prof. Felix Schacher, Jena) and parasitology (PD Simone Häberlein, Prof. Christoph Grevelding, Gießen), we apply functionalized hydrogels with mechanosensitive reporter functions for integration into organ-on-chip systems. These innovative materials enable the quantification of mechanical forces during parasite–host interactions and open new avenues for studying helminth infection biology and biophysics.
Project: Biomechanical forces and cellular responses in plathelminths and host cells | funded by the DFG within the Priority Program „Physics of Parasitism“ | 2025 – 2028
Autologous Stem Cell-Based Models for Personalized Drug Testing
We develop autologous iPSC-derived organ-on-chip systems that integrate isogenic adaptive and innate immune cells. These models reproduce patient-specific immune responses in defined tissue environments and provide a translational platform to study immune adaptation, drug safety, and immunotoxicity.
selected projects:
- Immune Safety Avatar: nonclinical mimicking of the immune system effects of immunomodulatory therapies (imSAVAR)| funded by the European Commission | 2020–2025
- Vascularised liver-on-chip model for personalised drug testing (VaskuChip) | funded by the Thüringer Aufbaubank – R&D Network Funding | 2024 – 2027
Who we are
Hier stellen wir Ihnen die engagierten Mitglieder und ihre Expertise vor.

Prof. Dr. Alexander S. Mosig
Head of Research Group
Professor of Biochemistry and Cell Biology, Non-Animal Methods in Infection and Inflammation Research

Dr. Nicole Engert
PostDoc
Project Lead Stem Cell-Based Gastrointestinal Model Development Norovirus Infection Models

Dr. Friedrich Becker
PostDoc
Project Lead Stem Cell-Based Liver Models for Assessment of Immonotoxicity

Dr. Mohamed Hassan
PostDoc
Project Lead Modelling Persistence of Staphylococcus aureus infections

MSc. Hristina Koceva
PhD Student
Development of iPSC-based Viral Resperatory Infection Models

MSC. Mona Amaratashani
PhD Student
Development of Respiratory Infections Models with Breathing Mechanics

Msc Maria Warschinke
PhD Student
Modelling Clostridioides difficile Infection in Stem Cell-Based Gastrointestinal Models

MSC Valentin Wegner
PhD Student
Investigating Role of the Microbiome on CAR T Cell Therapy in Gut-on-chip Models

MSc. Adrian Feile
PhD Student
Modelling Infection and Quorum Sensing of Vibrio cholerae in Gut-on-chip Models

Nora Mosig
Technician

Melanie Ulrich
Technician

Varis Malazezi
HiWi
Current Research Projects
Advancing 3R principles through human-relevant models.
funded by German Research foundation
Microbiome Interactions in the Respiratory and Gut Epithelium (MIRAGE)
MIRAGE aims to establish a novel human-based multi-organ-on-chip platform to investigate how nasal and gut microbiomes jointly shape respiratory infection outcomes with Staphylococcus aureus. The modular system will integrate immunocompetent nasal, bronchial, alveolar, and gut tissue models connected via microfluidic circuits mimicking airflow and systemic circulation. By modeling microbial metabolites and defined consortia, we will study how perturbations in the gut and nasal microbiota influence epithelial barrier function, immune competence, and infection severity along the respiratory axis. We aim to identify cross-compartmental microbiome traits and develop advanced in vitro models for translational infection research.
This work isfunded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy (EXC 2051, Project-ID 390713860)

Exploring the role of pathogen interactions in transitioning of ventilator-associated tracheobronchitis to pneumonia (VATOP)
We develop advanced alveolus- and bronchiole-on-chip models that replicate the human lung environment using iPSC-derived epithelial, endothelial, and immune cells. With these models, we investigate how commensals such as Staphylococcus epidermidis or Candida albicans influence infections with major pathogens like Pseudomonas aeruginosa and Staphylococcus aureus, and how host immune responses shape the transition from colonization to disease. By enabling long-term, immunocompetent air–liquid interface cultures with functional barrier and immune properties, our organ-on-chip systems provide human-relevant insight into respiratory infection mechanisms and support the 3R principle by reducing reliance on animal models.
Biomechanical forces and cellular responses in plathelminths and host cells
In the framework of the DFG Priority Programme „Physics of Parasitism“ (SPP 2332), we develop advanced worm-on-a-chip models to study the biomechanics of parasitic flatworms such as Schistosoma mansoni and Fasciola hepatica. Building on microfluidic biochips and mechano-responsive hydrogels, we recreate semi-natural flow conditions of blood vessels and bile ducts to quantify adhesion, locomotion, and reproduction of worms under physiologically relevant stress. By integrating host-derived liver sinusoid and cholangiocyte models into biochips, we investigate how parasites interact with human tissues, apply forces, and adapt their biology to the physical microenvironment. This provides quantitative insights into parasite–host biomechanics in a controlled chip setting and opens perspectives for translational applications in infection biology and drug discovery.

Probing cholera infection on a chip
Cholera, caused by the human-specific pathogen Vibrio cholerae, remains poorly understood due to the limitations of rodent models and conventional cell culture systems. In this project, we use a gut-on-a-chip model to recreate the interaction between V. cholerae and human intestinal cells and to investigate how bacterial quorum sensing influences colonization and biofilm formation. By combining advanced imaging and transcriptomic analyses with studies of quorum sensing mutants, we aim to unravel the molecular mechanisms driving infection. This human-relevant model provides a powerful platform to study cholera pathogenesis under physiologically realistic conditions.
This work isfunded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy (EXC 2051, Project-ID 390713860)
funded by the european COMMISSION
MPS@NOVA Hub: Mechanisms of chronic diseases and Host-Microbe Interactions with Advanced Microphysiological Systems and Pluripotent Stem Cell Technologies
MPS@NOVA brings together NOVA University of Lisbon (ITQB NOVA, NOVA Medical School), Fondazione Human Technopole (Italy), Berlin Institute for Medical Systems Biology at MDC (Germany), and University Hospital Jena (Germany). At UKJ, we contribute pioneering organ-on-chip infection and inflammation models to study host–microbe interactions and immune responses. Together, the consortium advances stem cell–based MPS, strengthens European excellence, and fosters translation toward clinical applications.
Inno4Vac – Human next generation mucosal models and assays to accelerate vaccine development
Inno4Vac project is an ambitious public-private partnership dedicated to accelerating vaccine development. It focuses on tackling scientific bottlenecks by leveraging cutting-edge computational modeling and artificial intelligence. The project aims to create a cloud-based platform for the in silico assessment of vaccine efficacy, and a simulation platform to guide bio-manufacturing processes. A key part of this effort involves developing novel cell-based human in vitro 3D models of the respiratory and gastrointestinal tracts to better simulate infections and predict immune protection. These models are crucial for enabling the early evaluation of vaccine efficacy and de-risking the development process.
Immune Safety Avatar: nonclinical mimicking of the immune system effects of immunomodulatory therapies
The Immune Safety Avatar is a collaborative initiative aimed at enhancing the safety and efficacy of immunomodulatory therapies. Funded by the Innovative Medicines Initiative (IMI), its main goal is to develop a platform for integrated nonclinical assessments. By creating fit-for-purpose models with high transferability, the project seeks to significantly improve the prediction of potential side effects, ultimately improving patient safety. This is achieved through a partnership of 28 international partners from academia, the private sector, regulators, and technology providers.
funded by Federal Ministry of Research, Technology and Space

Human infection model for Vibrio cholerae as an alternative to animal testing (VIBRO-3R)
The project aims to develop a human gut-on-a-chip model for Vibrio cholerae as a realistic alternative to animal testing. By replicating the biophysical, biochemical, and physiological aspects of the human gut in vitro, the model allows for a precise study of infection mechanisms, particularly in light of increasing antibiotic resistance and climate-change-related epidemics. Utilizing microfluidic technologies and AI-based image analysis, the project enables the quantification of V. cholerae biofilm formation under realistic conditions, thereby facilitating the development of new therapeutic approaches, such as phage therapies. The model is intended to be a standardized, regulatorily relevant testing platform for new drugs, with high transfer potential for other enteropathogenic bacteria.
Automated organ-on-chip platform for infection research (AutOoC)
Persistent lung infections caused by drug-resistant bacteria like MRSA are a growing medical crisis. Our project pioneers a unique Human Lung-on-Chip Model built from patient-derived stem cells, which accurately mimics the complex environment of the human lung. This allows us to study how these bacteria evade treatment by hiding inside cells and developing resistance. The platform enables us to test innovative diagnostic strategies and therapies, aiming to significantly improve the accuracy of preclinical tests and accelerate personalized medicine while reducing the need for animal testing. We are creating a new standard in infection research for a healthier, safer future.
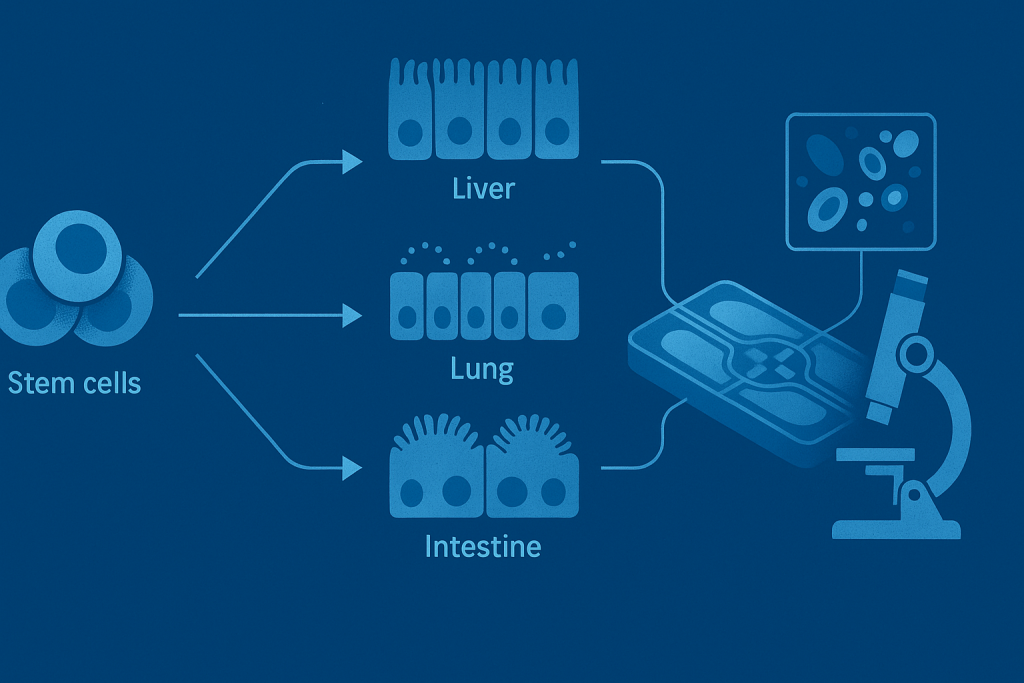
Profiling of host-response to life threatening infections (Leibniz Institute of Photonics in Infections)
The project aims to develop personalized infection models for organ-on-chip systems, offering a realistic in vitro alternative to animal testing. Using human induced pluripotent stem cells (iPSCs), we create self-structuring complex in vitro models that mimic the microarchitecture of human organs such as the lungs, liver, and gut. To facilitate detailed analysis, the project will also develop optical interfaces for these chips, enabling high-resolution microscopy to study infection mechanisms and measure pathogen loads across different cell types. Ultimately, this approach will help identify biomarkers, evaluate new drug candidates, and deepen our understanding of infections within a more physiologically relevant context, with the goal of establishing a new diagnostic platform for personalized medicine.
funded by the Federal Ministry of Economic Affairs and Energy
Immunocompatible microphysiological system with flexible collagen membrane to create physiological micro-environments for organ-on-chip models
The IMPROVE project is developing next-generation microphysiological systems for studying human respiratory infections. We are developing advanced lung-on-chip models that surpass simple cell cultures by incorporating three critical components: a flexible, bio-hybrid membrane made from human-derived collagen, a technology for multifactorial mechanical stimulation (mimicking breathing mechanics), and an immune system component. By creating a more realistic microenvironment, our models will provide an unprecedented platform for studying complex human disease mechanisms.
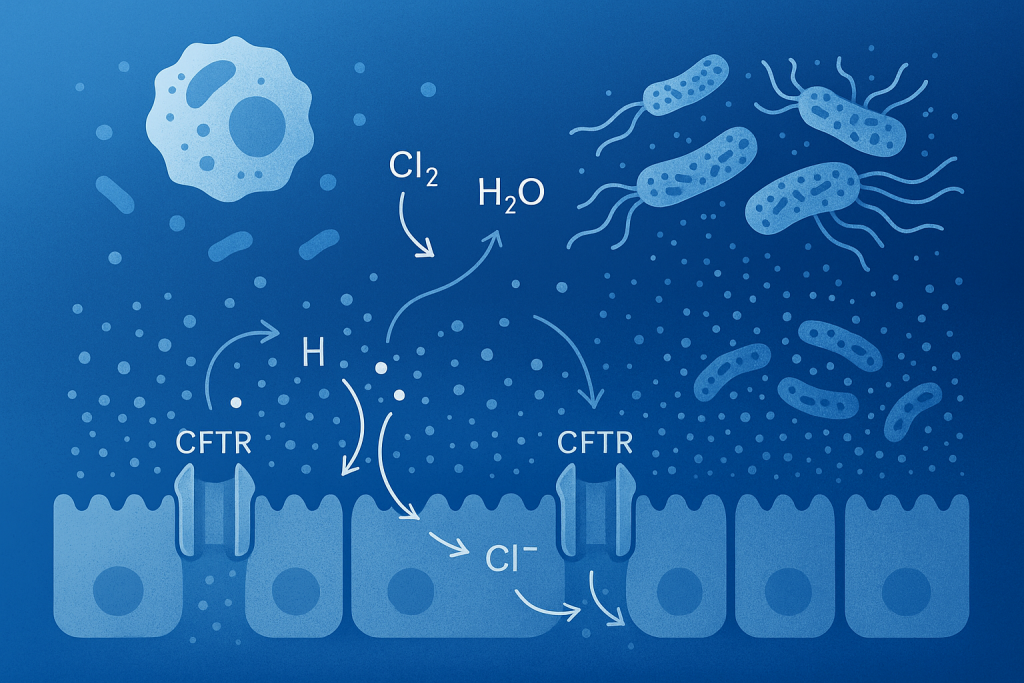
Modelling cystic fibrosis and associated infections in a bronchiole-on-chip model for drug testing
CF-on-chip project aims to model cystic fibrosis (CF) and its associated infections in a bronchiolus-on-chip system, providing a realistic alternative to animal testing. CF is a common, life-shortening genetic disease characterized by thickened mucus in the airways, leading to chronic infections often caused by Staphylococcus aureus. The project, a collaboration between Dynamic42 GmbH and the University Hospital Jena, will develop an isogenetic and immune-competent bronchiolus model from human stem cells. The new microfluidic platform will integrate optical interfaces and oxygen sensors to replicate the CF phenotype and test the effectiveness of new drugs and antibiotics. This approach seeks to improve the accuracy of preclinical tests and accelerate the development of more efficient therapies for this disease.
funded by the Thüringer Aufbaubank
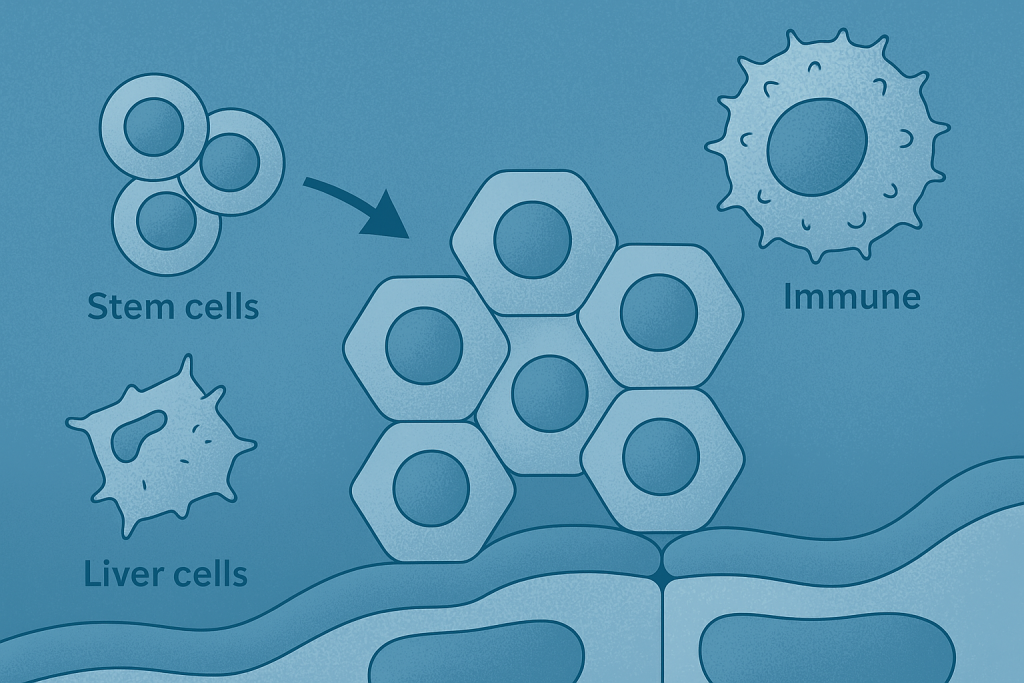
Vascularised liver-on-chip model for personalised drug testing
This VascuLiver project focuses on the development of a highly advanced, vascularized liver-on-chip model. Using human induced pluripotent stem cells (iPSCs), the goal is to create a complex, multi-cellular tissue that includes liver cells, blood vessel structures, and an immune component with circulating T-cells. This system is designed to provide a realistic platform for testing the safety and efficacy of new drugs. By replicating the complex interactions within a human liver, the model can be used to study drug-induced side effects, particularly those related to the immune system.

Vascularised liver-on-chip model for personalised drug testing
This VascuLiver project focuses on the development of a highly advanced, vascularized liver-on-chip model. Using human induced pluripotent stem cells (iPSCs), the goal is to create a complex, multi-cellular tissue that includes liver cells, blood vessel structures, and an immune component with circulating T-cells. This system is designed to provide a realistic platform for testing the safety and efficacy of new drugs. By replicating the complex interactions within a human liver, the model can be used to study drug-induced side effects, particularly those related to the immune system.
Publications
2025
Kapitan M., Niemiec M.J., Swidergall M., Millet N., Brandt P., Chowdhury E., Hoffmann F., Höring S., Lange A., Veleba M., Nietzsche S., Mosig A.S., Löffler B., Kline K., Vylkova S., Jacobsen I.D., „Synergistic cross-kingdom host cell damage with Candida albicans relies on Enterococcus faecalis cytolysin„, Proceedings of the National Academy of Sciences, accepted
Gheitasi R., Weiss D., Müller M.M., Sommer K., Roell D., Mosig A.S., Pletz M.W., Makarewicz O. (2025) „Staphylococcus aureus biofilm-associated component PNAG stimulates the secretion of the immunomodulatory chemokine CXCL10 via Dectin-1 signaling“ Communications Biology, Jul 31;8(1):1136. doi: 10.1038/s42003-025-08503-z,
Wegner, V. D., Feile, A., Alb, M., Hudecek, M., Hewitt, P., & Mosig, A. S. (2025). „Short-Chain Fatty Acids Modulate Anti-ROR1 CAR T-Cell Function and Exhaustion in an Intestinal Adenocarcinoma-on-Chip Model“ Advanced Healthcare Materials, e2405003. Advance online publication. https://doi.org/10.1002/adhm.202405003
Koceva, H., Amiratashani, M., Akbarimoghaddam, P., Hoffmann, B., Zhurgenbayeva, G., Gresnigt, M.S., Rossetto Marcelino, V., Eggeling, C., Figge, M.T., João Amorim, M., Mosig, A.S., (2025) „Deciphering Respiratory Viral Infections by Harnessing Organ-on-Chip Technology to Explore the Gut-Lung Axis„. Open Biology, Mar;15(3):240231. doi: 10.1098/rsob.240231. Epub 2025 Mar 5.
Kaden, T., Alonso-Román, R., Stallhofer, J., Gresnigt, M. S., Hube, B., & Mosig, A. S., (2025) „Leveraging Organ-on-Chip Models to Investigate Host-Microbiota Dynamics and Targeted Therapies for Inflammatory Bowel Disease“. Advanced Healthcare Materials, e2402756. Advance online publication. https://doi.org/10.1002/adhm.202402756
2024
Liu H., Yin G., Kohlhepp M.S., Schumacher F., Hundertmark J., Abdelwahab Hassan M., Heymann F., Puengel T., Kleuser B., Mosig A.S., Tacke F., Guillot A. (2024), „Dissecting pathomechanisms and therapeutic responses of steatotic liver disease using primary 2 mouse liver and blood cells in a liver-on-a-chip model„, Advanced Sciences, 11(30), e2403516. https://doi.org/10.1002/advs.202403516
Feile A., Wegner V., Mosig A.S. (2024), „Immunocompetent Intestine-on-Chip Model for Analyzing Gut Mucosal Immune Responses„, Journal of Visualized Experiments : JoVE, 10.3791/66603. https://doi.org/10.3791/66603
Koceva, H., Amiratashani, M., Rennert, K., & Mosig, A. S. (2024). „Immunocompetent Alveolus-on-Chip Model for Studying Alveolar Mucosal Immune Responses.“ Journal of Visualized Experiments : JoVE, (207), 10.3791/66602. https://doi.org/10.3791/66602
Ehle C., Iyer-Bierhoff A., Wu Y., Xing S., Kiehntopf M., Mosig A.S., Godmann M., Heinzel T. (2024), „Repression of HNF4A enables transcriptomic reprogramming during the hepatic acute-phase response„, Communications Biology, 7(1), 589. https://doi.org/10.1038/s42003-024-06288-1
Kaden, T.*, Alonso-Román R.*, Akbarimoghaddam P.*, Mosig A.S., Graf K., Raasch M., Hoffmann B., Figge M.T., Hube. B., Gresnigt M.S. (2024) „Modeling of intravenous caspofungin administration using an intestine-on-chip reveals altered Candida albicans microcolonies and pathogenicity„, Biomaterials, ahead of print, doi:10.1016/j.biomaterials.2024.122525 (* contributed equally)
Lucchettia M., Kehinde Oluwasegunb A., Grandmougina L, Jägera C., Perez-Escrivac P., Letellierc E., Mosig A.S., Wilmes P. (2024) „An organ-on-chip platform for simulating drug metabolism along the gut-liver axis„, Advanced Healthcare Materials, ahead of print, doi: 10.1002/adhm.202303943
Roth A., Tannert A., Ziller N., Eiserloh A., Göhrig B., Guliev RR., Gonzalez Vazquez MJ., Naumann M., Mosig AS., Stengel S., Heutelbeck ARR., Neugebauer U., (2024) „Quantification of polystyrene uptake by different cell lines using fluorescence microscopy and label-free visualization of intracellular polystyrene particles by Raman microspectroscopic imaging“, Cells, ahead of print, doi:10.3390/cells13050454
Kowalczuk K., Wegner V., Mosig AS.*, Schacher, F.H.*, (2024) „Tailoring the degradation time of polycationic PEG-based hydrogels towards dynamic cell culture matrices„, ACS Applied Bio Materials, ahead of print, doi:10.1021/acsabm.4c00057 (* shared senior authorship)
Kowalczuk K., Dasgupta A., Páez Larios F., Ulrich H.F., Wegner V., Brendel J.C., Eggeling C., Mosig A.S., Schacher F.H., (2024) „Self-degrading multifunctional PEG-based hydrogels – tailormade substrates for cell culture„, Macromolecular Bioscience, ahead of print, doi:10.1002/mabi.202300383
Kowalczuk, K., Mons, P. J., Ulrich, H. F., Wegner, V. D., Brendel, J. C., Mosig, A. S., & Schacher, F. H. (2024). Asymmetric Block Extension of Star-Shaped [PEG-SH]4 – toward Poly(dehydroalanine)-Functionalized PEG Hydrogels for Catch and Release of Charged Guest Molecules. Macromolecular Bioscience, e2300230. Advance online publication. https://doi.org/10.1002/mabi.202300230
Alonso-Roman R., Mosig A.S., Figge M.T., Papenfort K., Eggeling C., Schacher F.H., Hube B., Gresnig M.S. (2024), „Organ-on-chip models for infectious disease research„, Nature Microbiology, ahead of print, doi: 10.1038/s41564-024-01645-6.
2023
Fahrner, R., Gröger, M., Settmacher, U., & Mosig, A. S. (2023). Functional integration of natural killer cells in a microfluidically perfused liver on-a-chip model. BMC Research Notes, 16(1), 285. https://doi.org/10.1186/s13104-023-06575-w
Kaden, T., Graf, K., Rennert, K., Li, R., Mosig, A. S., & Raasch, M. (2023). Evaluation of drug-induced liver toxicity of trovafloxacin and levofloxacin in a human microphysiological liver model. Scientific Reports, 13(1), 13338. https://doi.org/10.1038/s41598-023-40004-z
Ney, L. M., Wipplinger, M., Grossmann, M., Engert, N., Wegner, V. D., & Mosig, A. S. (2023). Short chain fatty acids: key regulators of the local and systemic immune response in inflammatory diseases and infections. Open Biology, 13(3), 230014. https://doi.org/10.1098/rsob.230014
2022
Siwczak, F., Cseresnyes, Z., Hassan, M. I. A., Aina, K. O., Carlstedt, S., Sigmund, A., Groger, M., Surewaard, B. G. J., Werz, O., Figge, M. T., Tuchscherr, L., Loffler, B., & Mosig, A. S. (2022). Human macrophage polarization determines bacterial persistence of Staphylococcus aureus in a liver-on-chip-based infection model. Biomaterials, 287, 121632. Advance online publication. https://doi.org/10.1016/j.biomaterials.2022.121632
Hoang, T. N. M., Cseresnyés, Z., Hartung, S., Blickensdorf, M., Saffer, C., Rennert, K., Mosig, A. S., von Lilienfeld-Toal, M., & Figge, M. T. (2022). Invasive aspergillosis-on-chip: A quantitative treatment study of human Aspergillus fumigatus infection. Biomaterials, 283, 121420. https://doi.org/10.1016/j.biomaterials.2022.121420
Ehsani, M., Bartsch, S., Rasa, S. M. M., Dittmann, J., Pungsrinont, T., Neubert, L., Huettner, S. S., Kotolloshi, R., Schindler, K., Ahmad, A., Mosig, A. S., Adam, L., Ori, A., Neri, F., Berndt, A., Grimm, M. O., & Baniahmad, A. (2022). The natural compound atraric acid suppresses androgen-regulated neo-angiogenesis of castration-resistant prostate cancer through angiopoietin 2. Oncogene, 41(23), 3263–3277. https://doi.org/10.1038/s41388-022-02333-7
Shroff, T., Aina, K., Maass, C., Cipriano, M., Lambrecht, J., Tacke, F., Mosig, A.S., & Loskill, P. (2022). Studying metabolism with multi-organ chips: new tools for disease modelling, pharmacokinetics and pharmacodynamics. Open Biology, 12(3), 210333. https://doi.org/10.1098/rsob.210333
2021
Neukirch, K., Alsabil, K., Dinh, C. P., Bilancia, R., Raasch, M., Ville, A., Cerqua, I., Viault, G., Bréard, D., Pace, S., Temml, V., Brunner, E., Jordan, P. M., Marques, M. C., Loeser, K., Gollowitzer, A., Permann, S., Gerstmeier, J., Lorkowski, S., Stuppner, H, Garscha U, Rodrigues T, Bernardes GJL, Schuster D, Séraphin D, Richomme P, Rossi A, Mosig A. S., Roviezzo F, Werz O, Helesbeux JJ, Koeberle, A. (2021). Exploration of Long-Chain Vitamin E Metabolites for the Discovery of a Highly Potent, Orally Effective, and Metabolically Stable 5-LOX Inhibitor that Limits Inflammation. Journal of Medicinal Chemistry, 64(15), 11496–11526. https://doi.org/10.1021/acs.jmedchem.1c00806 –
Deinhardt-Emmer, S., Böttcher, S., Häring, C., Giebeler, L., Henke, A., Zell, R., Jungwirth, J., Jordan, P. M., Werz, O., Hornung, F., Brandt, C., Marquet, M., Mosig, A. S., Pletz, M. W., Schacke, M., Rödel, J., Heller, R., Nietzsche, S., Löffler, B., & Ehrhardt, C. (2021). SARS-CoV-2 causes severe epithelial inflammation and barrier the epithelial barrier. Journal of Virology, 95(10), e00110-21. https://doi.org/10.1128/JVI.00110-21
Heinze, K., Hölzer, M., Ungelenk, M., Gerth, M., Thomale, J., Heller, R., Morden, C. R., McManus, K. J., Mosig, A.S., Dürst, M., Runnebaum, I. B., & Häfner, N. (2021). RUNX3 Transcript Variants Have Distinct Roles in Ovarian Carcinoma and Differently Influence Platinum Sensitivity and Angiogenesis. Cancers, 13(3), 476. https://doi.org/10.3390/cancers13030476
Gresing LJ, Radon P, Friedrich RP, Zahn D, Raasch M, Mosig AS, Dutz S, Alexiou C, Wiekhorst F, Hochhaus A, Clement JH; (2021) Negatively charged magnetic nanoparticles pass the blood-placenta barrier under continuous flow conditions in a time-dependent manner. Journal of Magnetism and Magnetic Materials, 521(2):167535, https://doi.org/10.1016/j.jmmm.2020.167535 72
Lucchetti, M., Kaminska, M., Oluwasegun, A. K., Mosig, A. S., & Wilmes, P. (2021). Emulating the gut-liver axis: Dissecting the microbiome’s effect on drug metabolism using multiorgan-on-chip models. Current Opinion in Endocrine and Metabolic Research, 18, 94–101. https://doi.org/10.1016/j.coemr.2021.03.003
Siwczak, F., Loffet, E., Kaminska, M., Koceva, H., Mahe, M. M., & Mosig, A. S. (2021). Intestinal Stem Cell-on-Chip to Study Human Host-Microbiota Interaction. Frontiers in Immunology, 12, 798552. https://doi.org/10.3389/fimmu.2021.798552
Last, A., Maurer, M., Mosig, A.S., S Gresnigt, M., & Hube, B. (2021). In vitro infection models to study fungal-host interactions. FEMS Microbiology Reviews, 45(5), fuab005. https://doi.org/10.1093/femsre/fuab005.
2020
Diederich B., Lachmann R., Carlstedt S., Marsikova B., Wang H., Uwurukundu X., Mosig AS., Heintzmann R.; (2020) UC2 – A Versatile and Customizable low-cost 3D-printed Optical Open-Standard for microscopic imaging. Nature Communications, 11(1):5979
Tansi, F. L., Rüger, R., Böhm, C., Steiniger, F., Raasch, M., Mosig, A. S., Kontermann, R. E., Teichgräber, U. K., Fahr, A., & Hilger, I. (2020). Rapid Target Binding and Cargo Release of Activatable Liposomes Bearing HER2 and FAP Single-Chain Antibody Fragments Reveal Potentials for Image-Guided Delivery to Tumors. Pharmaceutics, 12(10), 972. https://doi.org/10.3390/pharmaceutics12100972
Schicke, E., Cseresnyés, Z., Rennert, K., Vau, V., Haupt, K. F., Hornung, F., Nietzsche, S., Swiczak, F., Schmidtke, M., Glück, B., Koch, M., Schacke, M., Heller, R., Mosig, A. S., Figge, M. T., Ehrhardt, C., Löffler, B., & Deinhardt-Emmer, S. (2020). Staphylococcus aureus Lung Infection Results in Down-Regulation of Surfactant Protein-A Mainly Caused by Pro-Inflammatory Macrophages. Microorganisms, 8(4), 577. https://doi.org/10.3390/microorganisms8040577
Spengler K., Kryeziu N., Große S., Mosig AS., Heller R.; (2020) VEGF Triggers Transient Induction of Autophagy in Endothelial Cells via AMPKα1. Cells 9(3), 687.
Deinhardt-Emmer, S., Rennert, K., Schicke, E., Cseresnyés, Z., Windolph, M., Nietzsche, S., Heller, R., Siwczak, F., Haupt, K. F., Carlstedt, S., Schacke, M., Figge, M. T., Ehrhardt, C., Löffler, B., & Mosig, A. S. (2020). Co-infection with Staphylococcus aureus after primary influenza virus infection leads to damage of the endothelium in a human alveolus-on-a-chip model. Biofabrication, 12(2), 025012. https://doi.org/10.1088/1758-5090/ab7073
Schubert, M., Becher, S., Wallert, M., Maeß, M. B., Abhari, M., Rennert, K., Mosig, A. S., Große, S., Heller, R., Grün, M., & Lorkowski, S. (2020). The Peroxisome Proliferator-Activated Receptor (PPAR)-γ Antagonist 2-Chloro-5-Nitro-N-Phenylbenzamide (GW9662) Triggers Perilipin 2 Expression via PPARδ and Induces Lipogenesis and Triglyceride Accumulation in Human THP-1 Macrophages. Molecular Pharmacology, 97(3), 212–225. https://doi.org/10.1124/mol.119.117887
2019
Graf, K., Last, A., Gratz, R., Allert, S., Linde, S., Westermann, M., Gröger, M., Mosig, A. S., Gresnigt, M. S., & Hube, B. (2019). Keeping Candidacommensal: how lactobacilli antagonize pathogenicity of Candida albicans in an in vitro gut model. Disease models & mechanisms, 12(9), dmm039719. https://doi.org/10.1242/dmm.039719
Maurer, M., Gresnigt, M. S., Last, A., Wollny, T., Berlinghof, F., Pospich, R., Cseresnyes, Z., Medyukhina, A., Graf, K., Gröger, M., Raasch, M., Siwczak, F., Nietzsche, S., Jacobsen, I. D., Figge, M. T., Hube, B., Huber, O., & Mosig, A. S. (2019). A three-dimensional immunocompetent intestine-on-chip model as in vitro platform for functional and microbial interaction studies. Biomaterials, 220, 119396. https://doi.org/10.1016/j.biomaterials.2019.119396
Blaurock-Möller, N., Gröger, M., Siwczak, F., Dinger, J., Schmerler, D., Mosig, A. S.*, & Kiehntopf, M*. (2019). CAAP48, a New Sepsis Biomarker, Induces Hepatic Dysfunction in an in vitro Liver-on-Chip Model. Frontiers in Immunology, 10, 273. https://doi.org/10.3389/fimmu.2019.00273
Raasch, M., Fritsche, E., Kurtz, A., Bauer, M., & Mosig, A. S. (2019). Microphysiological systems meet hiPSC technology – New tools for disease modeling of liver infections in basic research and drug development. Advanced Drug Delivery Reviews, 140, 51–67. https://doi.org/10.1016/j.addr.2018.06.008
2018
Ryabchykov O., Bräutigam K., Galler K., Neugebauer U., Mosig A.S., Bocklitz T., Popp J. (2018) Characterization of the unique human liver cell line HepaRG by Raman microspectroscopy. Journal of Raman Spectroscopy, June; 49(6): 935-942, DOI: 10.1002/jrs.5392
Leonhardt, J., Große, S., Marx, C., Siwczak, F., Stengel, S., Bruns, T., Bauer, R., Kiehntopf, M., Williams, D. L., Wang, Z. Q., Mosig, A. S., Weis, S., Bauer, M., & Heller, R. (2018). Candida albicans β-Glucan Differentiates Human Monocytes Into a Specific Subset of Macrophages. Frontiers in Immunology, 9, 2818. https://doi.org/10.3389/fimmu.2018.02818
Pein H, Ville A, Pace S, Temml V, Garscha U, Raasch M, Alsabil K, Viault G, Dinh CP, Guilet D, Troisi F, Neukirch K, König S, Bilancia R, Waltenberger B, Stuppner H, Wallert M, Lorkowski S, Weinigel C, Rummler S, Birringer M, Roviezzo F, Sautebin L, Helesbeux JJ, Séraphin D, Mosig AS, Schuster D, Rossi A, Richomme P, Werz O, Koeberle A. (2018). Endogenous metabolites of vitamin E limit inflammation by targeting 5-lipoxygenase. Nature Communications, 9(1), 3834. https://doi.org/10.1038/s41467-018-06158-5
Ryabchykov O., Bräutigam K., Galler K., Neugebauer U., Mosig AS., Bocklitz T., Popp J; (2018) Characterization of the unique human liver cell line HepaRG by Raman microspectroscopy. Journal of Raman Spectroscopy, , 49, 935-942
Heinze, K., Kritsch, D., Mosig, A. S., Dürst, M., Häfner, N., & Runnebaum, I. B. (2018). Functional Analyses of RUNX3 and CaMKIINα in Ovarian Cancer Cell Lines Reveal Tumor-Suppressive Functions for CaMKIINα and Dichotomous Roles for RUNX3 Transcript Variants. International Journal of Molecular Sciences, 19(1), 253. https://doi.org/10.3390/ijms19010253
Gröger, M., Dinger, J., Kiehntopf, M., Peters, F. T., Rauen, U., & Mosig, A. S. (2018). Preservation of Cell Structure, Metabolism, and Biotransformation Activity of Liver-On-Chip Organ Models by Hypothermic Storage. Advanced Healthcare Materials, 7(2), 10.1002/adhm . 201700616. https://doi.org/10.1002/adhm.201700616
2017
Fahrner, R., Möller, A., Press, A. T., Kortgen, A., Kiehntopf, M., Rauchfuss, F., Settmacher, U., & Mosig, A. S. (2017). Short-term treatment with taurolidine is associated with liver injury. BMC Pharmacology & Toxicology, 18(1), 61. https://doi.org/10.1186/s40360-017-0168-z
Rennert, K., Nitschke, M., Wallert, M., Keune, N., Raasch, M., Lorkowski, S., & Mosig, A. S. (2017). Thermo-responsive cell culture carrier: Effects on macrophage functionality and detachment efficiency. Journal of Tissue Engineering, 8, 2041731417726428. https://doi.org/10.1177/2041731417726428
Kritsch, D., Hoffmann, F., Steinbach, D., Jansen, L., Mary Photini, S., Gajda, M., Mosig, A. S., Sonnemann, J., Peters, S., Melnikova, M., Thomale, J., Dürst, M., Runnebaum, I. B., & Häfner, N. (2017). Tribbles 2 mediates cisplatin sensitivity and DNA damage response in epithelial ovarian cancer. International Journal of Cancer, 141(8), 1600–1614. https://doi.org/10.1002/ijc.30860
Mann, A., Pelz, T., Rennert, K., Mosig, A.S., Decker, M., & Lupp, A. (2017). Evaluation of HepaRG cells for the assessment of indirect drug-induced hepatotoxicity using INH as a model substance. Human Cell, 30(4), 267–278. https://doi.org/10.1007/s13577-017-0175-9
Thomas, L., Rao, Z., Gerstmeier, J., Raasch, M., Weinigel, C., Rummler, S., Menche, D., Müller, R., Pergola, C., Mosig, A.S., & Werz, O. (2017). Selective upregulation of TNFα expression in classically-activated human monocyte-derived macrophages (M1) through pharmacological interference with V-ATPase. Biochemical Pharmacology, 130, 71–82. https://doi.org/10.1016/j.bcp.2017.02.004
Pergola, C., Schubert, K., Pace, S., Ziereisen, J., Nikels, F., Scherer, O., Hüttel, S., Zahler, S., Vollmar, A. M., Weinigel, C., Rummler, S., Müller, R., Raasch, M., Mosig, A.S., Koeberle, A., & Werz, O. (2017). Modulation of actin dynamics as potential macrophage subtype-targeting anti-tumour strategy. Scientific Reports, 7, 41434. https://doi.org/10.1038/srep41434
Schüler, R., Osterhoff, M. A., Frahnow, T., Seltmann, A. C., Busjahn, A., Kabisch, S., Xu, L., Mosig, A. S., Spranger, J., Möhlig, M., Hornemann, S., Kruse, M., & Pfeiffer, A. F. (2017). High-Saturated-Fat Diet Increases Circulating Angiotensin-Converting Enzyme, Which Is Enhanced by the rs4343 Polymorphism Defining Persons at Risk of Nutrient-Dependent Increases of Blood Pressure. Journal of the American Heart Association, 6(1), e004465. https://doi.org/10.1161/JAHA.116.004465
Gröger, M., Lange, M., Rennert, K., Kaschowitz, T., Plettenberg, H., Hoffmann, M., & Mosig, A. S. (2017). Novel approach for the prediction of cell densities and viability in standardized translucent cell culture biochips with near infrared spectroscopy. Engineering in Life Sciences, 17(5), 585–593. https://doi.org/10.1002/elsc.201600162
2016
Englert, C., Trützschler, A. K., Raasch, M., Bus, T., Borchers, P., Mosig, A. S.*, Traeger, A.*, & Schubert, U. S.* (2016). Crossing the blood-brain barrier: Glutathione-conjugated poly(ethylene imine) for gene delivery. Journal of Controlled Release, 241, 1–14. https://doi.org/10.1016/j.jconrel.2016.08.039
Raasch, M., Rennert, K., Jahn, T., Gärtner, C., Schönfelder, G., Huber, O., Seiler, A. E., & Mosig A. S. (2016). Organ-on-chip models: new opportunities for biomedical research. Future Science OA, 3(2), FSO130. https://doi.org/10.4155/fsoa-2016-0038
(2016). An integrative microfluidically supported in vitro model of an endothelial barrier combined with cortical spheroids simulates effects of neuroinflammation in neocortex development. Biomicrofluidics, 10(4), 044102. https://doi.org/10.1063/1.4955184
Rennert, K., Heisig, K., Groeger, M., Wallert, M., Funke, H., Lorkowski, S., Huber, O., & Mosig, A. S. (2016). Recruitment of CD16(+) monocytes to endothelial cells in response to LPS-treatment and concomitant TNF release is regulated by CX3CR1 and interfered by soluble fractalkine. Cytokine, 83, 41–52. https://doi.org/10.1016/j.cyto.2016.03.017
Gröger, M., Rennert, K., Giszas, B., Weiß, E., Dinger, J., Funke, H., Kiehntopf, M., Peters, F. T., Lupp, A., Bauer, M., Claus, R. A., Huber, O., & Mosig, A. S. (2016). Monocyte-induced recovery of inflammation-associated hepatocellular dysfunction in a biochip-based human liver model. Scientific Reports, 6, 21868. https://doi.org/10.1038/srep21868
Rennert, K., Otto, P., Funke, H., Huber, O., Tomaso, H., & Mosig, A. S. (2016). A human macrophage-hepatocyte co-culture model for comparative studies of infection and replication of Francisella tularensis LVS strain and subspecies holarctica and mediasiatica. BMC Microbiology, 16, 2. https://doi.org/10.1186/s12866-015-0621-3
Press, A. T., Ungelenk, L., Rinkenauer, A. C., Gröger, M., Lehmann, F., Mosig, A.S., Schubert, U. S., Clemens, M. G., & Bauer, M. (2016). A new fluorescent dye for cell tracing and mitochondrial imaging in vitro and in vivo. Journal of Biophotonics, 9(9), 888–900. https://doi.org/10.1002/jbio.201500190
Mosig A. S. (2016). Organ-on-chip models: new opportunities for biomedical research. Future Science OA, 3(2), FSO130. https://doi.org/10.4155/fsoa-2016-0038
2015
Rennert, K., Steinborn, S., Gröger, M., Ungerböck, B., Jank, A. M., Ehgartner, J., Nietzsche, S., Dinger, J., Kiehntopf, M., Funke, H., Peters, F. T., Lupp, A., Gärtner, C., Mayr, T., Bauer, M., Huber, O., & Mosig, A. S. (2015). A microfluidically perfused three dimensional human liver model. Biomaterials, 71, 119–131. https://doi.org/10.1016/j.biomaterials.2015.08.043
Gögebakan, Ö., Osterhoff, M. A., Schüler, R., Pivovarova, O., Kruse, M., Seltmann, A. C., Mosig, A. S., Rudovich, N., Nauck, M., & Pfeiffer, A. F. (2015). GIP increases adipose tissue expression and blood levels of MCP-1 in humans and links high energy diets to inflammation: a randomised trial. Diabetologia, 58(8), 1759–1768. https://doi.org/10.1007/s00125-015-3618-4
Rinkenauer, A. C., Press, A. T., Raasch, M., Pietsch, C., Schweizer, S., Schwörer, S., Rudolph, K. L., Mosig, A.S., Bauer, M., Traeger, A., & Schubert, U. S. (2015). Comparison of the uptake of methacrylate-based nanoparticles in static and dynamic in vitro systems as well as in vivo. Journal of Controlled Release, 216, 158–168. https://doi.org/10.1016/j.jconrel.2015.08.008
Maeß M., Keller A.A., Rennert K., Mosig A.S., Lorkowski S.; Optimization of the transfection of human THP-1 macrophages by application of Nunc UpCell technology. Analytical Biochemistry, 2015, 479:40-42
Raasch, M., Rennert, K., Jahn, T., Peters, S., Henkel, T., Huber, O., Schulz, I., Becker, H., Lorkowski, S., Funke, H., & Mosig, A.S. (2015). Microfluidically supported biochip design for culture of endothelial cell layers with improved perfusion conditions. Biofabrication, 7(1), 015013. https://doi.org/10.1088/1758-5090/7/1/015013
Press, A. T., Traeger, A., Pietsch, C., Mosig, A.S., Wagner, M., Clemens, M. G., Jbeily, N., Koch, N., Gottschaldt, M., Bézière, N., Ermolayev, V., Ntziachristos, V., Popp, J., Kessels, M. M., Qualmann, B., Schubert, U. S., & Bauer, M. (2015). Cell type-specific delivery of short interfering RNAs by dye-functionalised theranostic nanoparticles. Nature Communications, 5, 5565. https://doi.org/10.1038/ncomms6565
Get in Touch
If you have questions or feedback we love to hear from you!
Find us here: https://maps.app.goo.gl/fb2ae7gSykwx2XZg7

